
DNA nanotechnology is a rapidly growing field of science that holds great promise for creating nanodevices capable of programmable transport and delivery of molecular cargoes, high-precision molecular sensing, nanomanufacturing, molecular computing, and countless other applications. However, for these applications to become a reality, the devices need to be rapidly actuated, precisely assembled into macroscopic arrays, and interfaced with inorganic and biological systems. Our lab uses theory, computations, and machine learning, of late, often in close collaboration with experimentalists, to address many of these challenges. We use methods grounded in statistical physics to account for thermal fluctuations and treat the devices at different resolutions to address their dynamics at various scales ranging from base pairing interactions to conformational transitions to mesoscopic diffusion and self-assembly (Fig. 1).
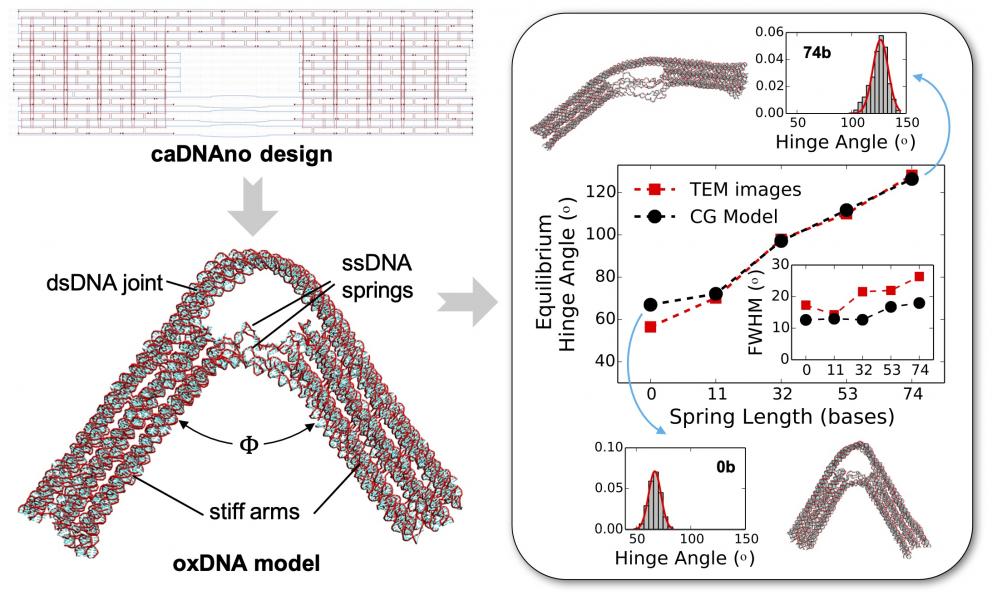
At the device level, we seek to understand and predict the equilibrium conformation and dynamics of compliant DNA nanodevices designed for force-sensing and nanorobotics applications. Our results show that coarse-grained models such as oxDNA can accurately reproduce the experimentally measured conformations of such devices (Fig. 2). More recently, we are developing models and simulation approaches to study biofunctionalized DNA origamis. In one collaborative project, we are using simulations to design and study the conformation of polynucleotide-brush functionalized DNA origami structures with enhanced stability against aggregation and enzymatic degradation. In another project, we are designing DNA origami platforms functionalized with various types of biomolecules for chromatin targeting, visualization, and engineering.
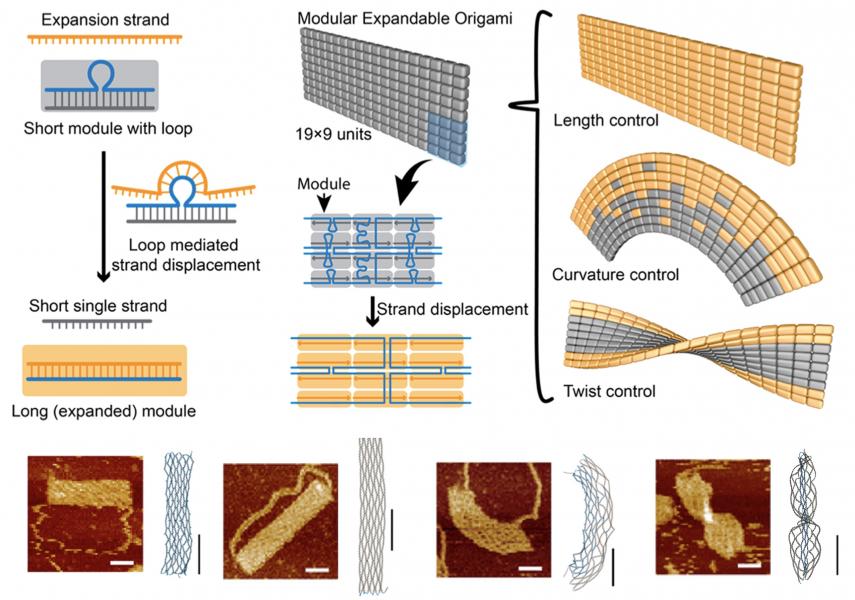
We have also been working closely with experimental groups to develop rapid and noninvasive strategies for actuating DNA devices. In one approach, we introduced multiple, weakly complementary pairs of single-stranded DNA overhangs to components of the structure, and by triggering the hybridization or dissociation of the overhangs via changes in solution ionic conditions we were able to drive structural transitions in the structure. By developing statistical mechanics models and efficient free energy calculation methods, we were able to model the actuation responses and provide design rules for tuning the actuation behavior. In another approach, we devised a modular design strategy that enables DNA structures to undergo multiple shape transformations based on the modular units being activated in the structure (Fig. 3). Using coarse-grained simulations, we were able to recapitulate and uncover the details of the observed shape transformations.

On method development, we have devised a new mesoscopic model of DNA with a switchable force field that can capture the folding process of DNA origamis, from start to finish. This has allowed us to study for the first time the kinetics and pathway of origami folding under various conditions as well as the role of scaffold routing and staple design on assembly yield. Other instances of methods we have developed include a software application for designing complex freeform DNA origami structures (Fig. 4) and a force-balance approach for rapidly predicting equilibrium conformations of multicomponent DNA structures based on force-deformation characteristics of the component connections.
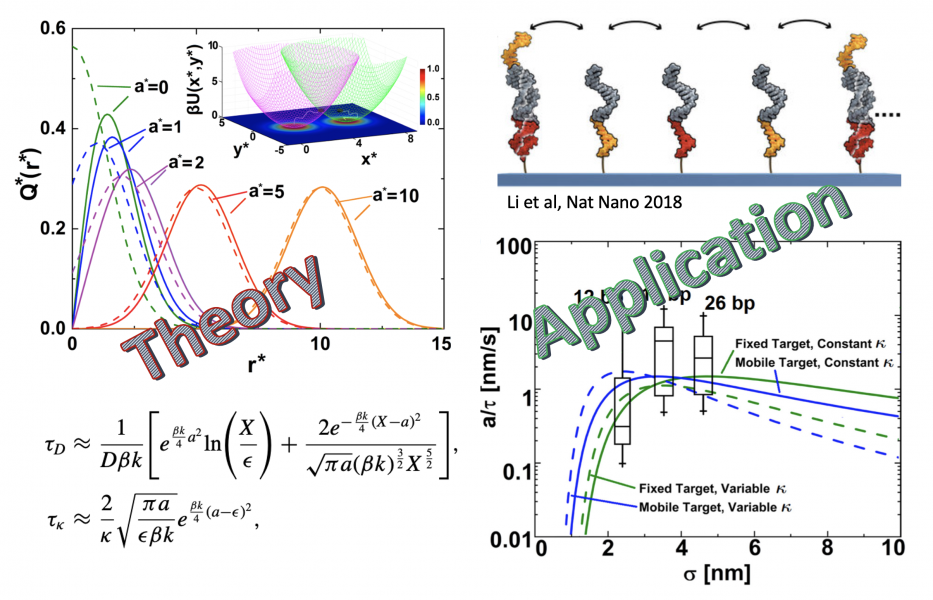
At the assembly level, we are developing strategies to assemble 1D and 2D arrays of such devices for communicating the sensed signals and magnifying the response by triggering collective behaviors like phase transitions. Simultaneously, we are developing theoretical and mesoscopic models to predict both the rate of signal communication across such devices and their order-disorder transitions. For instance, we recently developed an analytical theory for the binding time of field-confined Brownian particles and applied it to optimize cargo transport rates by DNA walkers (Fig. 5). In other projects, we are devising surface-mediated strategies to minimize defect formation during assembly of DNA tiles and polymer functionalization strategies to direct the placement of DNA structures on surfaces with precise positions and orientations. We are also developing novel machine learning strategies for the inverse design of patchy origami building blocks that would self-assemble into interesting superlattices.